よく実験結果の可視化などでSeabornを使用するので、使用方法をまとめておきます。随時追加していきます。
目次
Distplot(カーネル密度推定付き)
import pandas as pd
import numpy as np
import random
import seaborn as sns
import matplotlib.pyplot as plt
fig = plt.figure()
sns.set_style("darkgrid")
plt.xlabel('column_n')
n = [int(i) for i in range(0,1000)]
d = pd.DataFrame(
data = np.array([[random.randint(1,100) for j in n] for i in n]),
index = [f"r{i}" for i in n],
columns = [f"c{i}" for i in n]
)
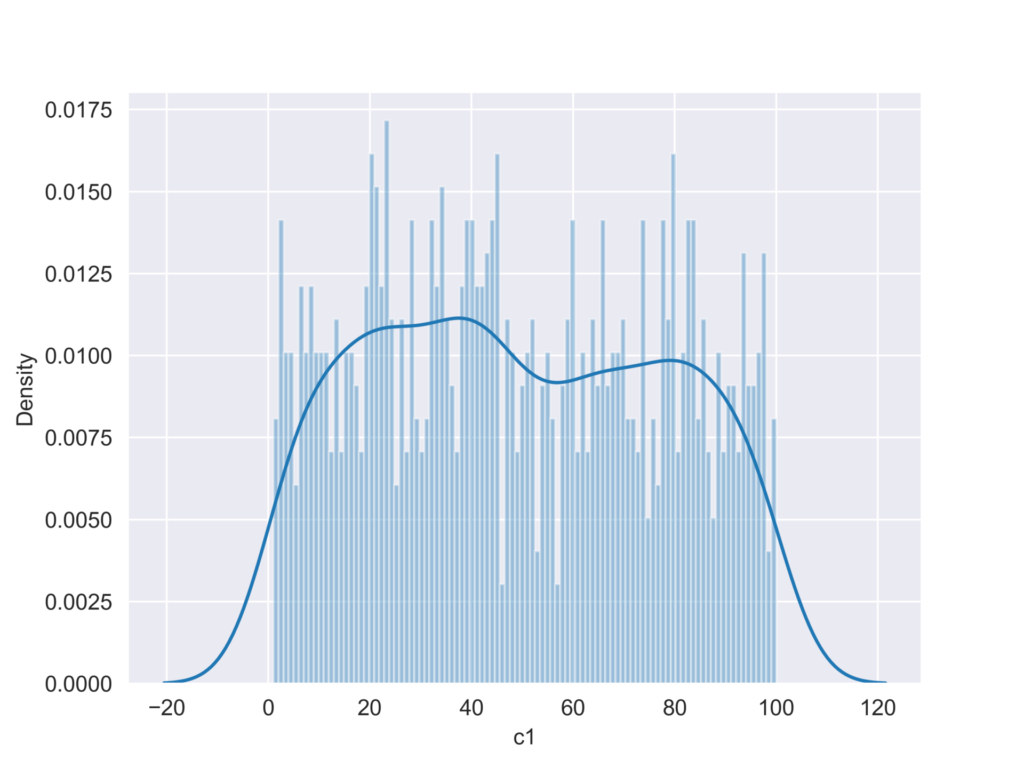
#distplot
sns.distplot(d["c1"],bins = 100,kde=True)
fig.savefig("fig.png",dpi = 500)
出力結果
Jointplot
import pandas as pd
import numpy as np
import random
import seaborn as sns
import matplotlib.pyplot as plt
plt.figure()
sns.set_style("darkgrid")
#deep, muted, pastel, bright, dark, colorblind
sns.set_palette("deep", n_colors=None, desat=None, color_codes=False)
n = [int(i) for i in range(0,1000)]
d = pd.DataFrame(
data = np.array([[random.randint(1,100) for j in n] for i in n]),
index = [f"r{i}" for i in n],
columns = [f"c{i}" for i in n]
)
#jointplot
#markers -> “.”,“,”,“o”,“*” etc.
sns.jointplot(x=d["c1"],y=d["c2"],data = d,s = 50, marker = ".")
plt.savefig("fig1.png",dpi = 700)
出力結果
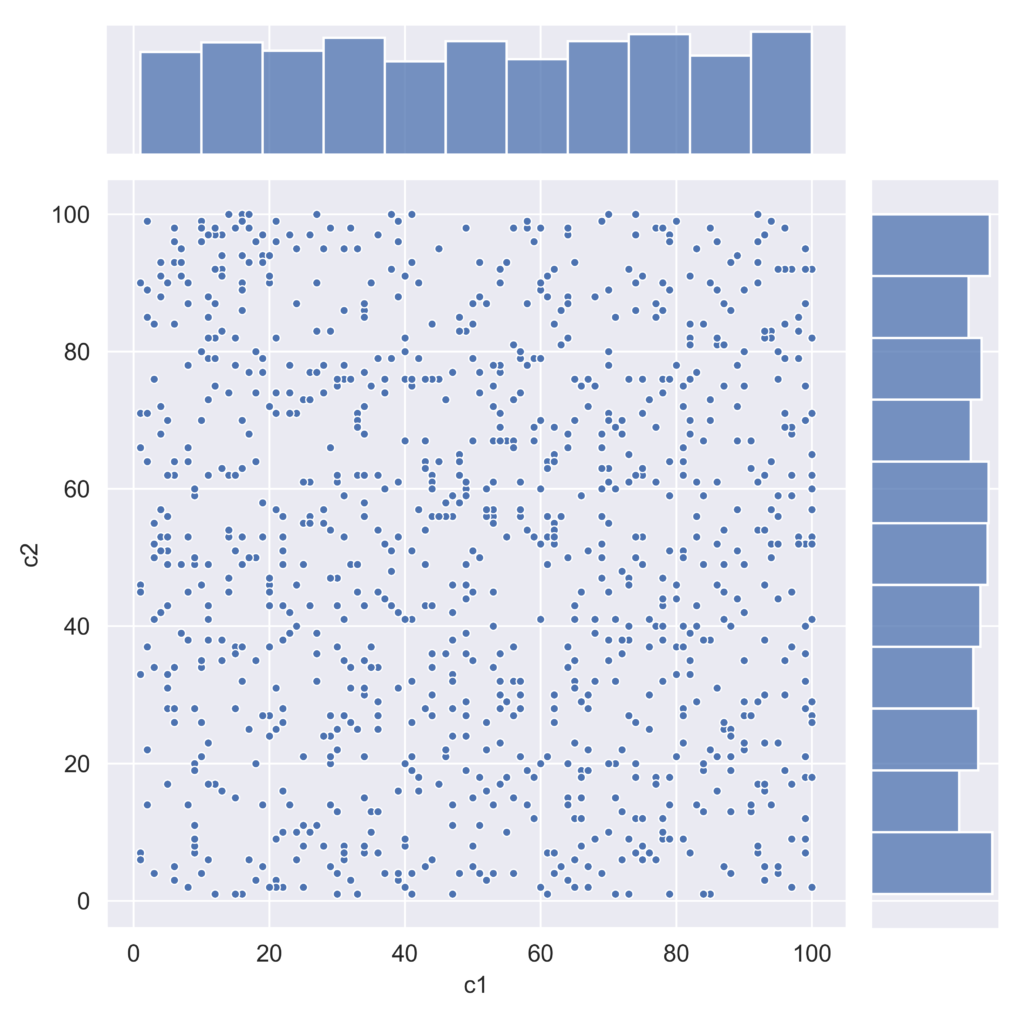
一様分布なので相関はありません。
Joint+線形回帰
#jointplot
#markers -> “.”,“,”,“o”,“*” etc.
sns.jointplot(x=d["c1"],y=d["c2"],data = d,kind="reg")
plt.savefig("fig1.png",dpi = 700)
これもランダムプロットのため面白みはありません。
実践
CDA測定結果のプロット
distplotを用いて、粒子サイズの分布を可視化してみます。この時、rugにより正規化しておきます。
import pandas as pd
import numpy as np
import random
import seaborn as sns
import matplotlib.pyplot as plt
fig = plt.figure()
sns.set_style("darkgrid")
plt.xlabel('column_n')
with open('6.csv') as f:
l = f.readlines()[6:]
df = [[(float(i.split(",")[1].split(' ')[0][1:]) + float(i.split(",")[1].split(' ')[2][:-1]))/2,int(i.split(",")[6][1])] for i in l[1:]]
df = pd.DataFrame(
data = np.array(df),
index = [f"r{i+1}" for i in range(len(df))],
columns = ["num","size"]
)
#distplot
sns.distplot(df["num"],bins = 15,kde=True,rug=False)
fig.savefig("fig.png",dpi = 500)実行結果
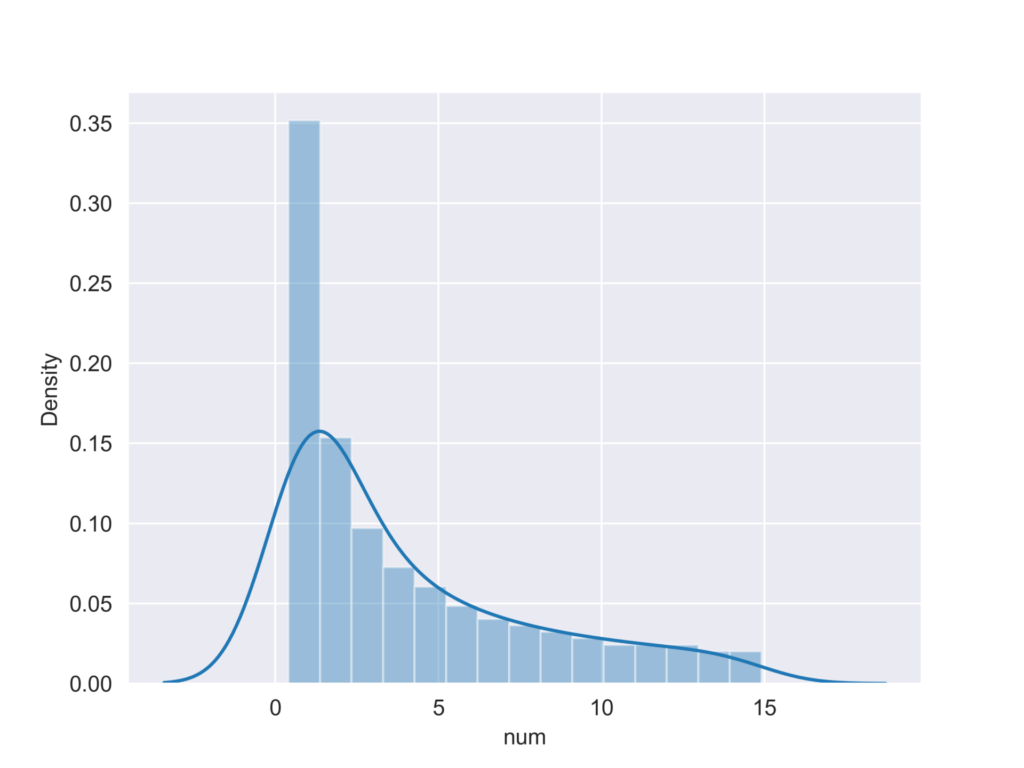
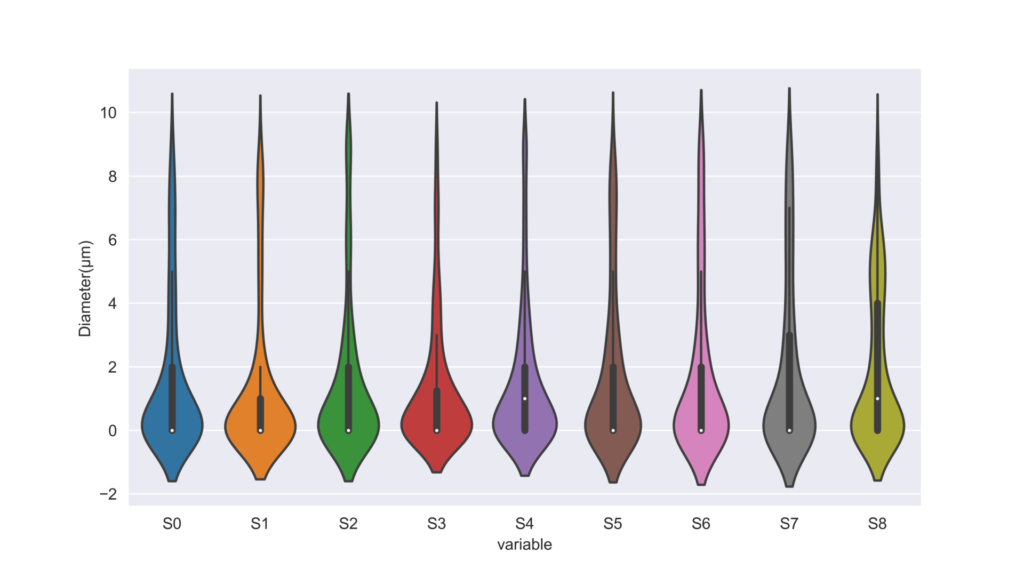
粒子サイズ分布の比較
複数系列での分布を比較できるバイオリンプロットを使用します。
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
def extract(n):
with open(f'dataset-2/{n}.csv',encoding='utf-8') as f:
l = f.readlines()[6:]
df = [[(float(i.split(",")[1].split(' ')[0][1:]) + float(i.split(",")[1].split(' ')[2][:-1]))/2,int(i.split(",")[6][1])] for i in l[1:]]
df = pd.DataFrame(
data = np.array(df),
index = [f"r{i+1}" for i in range(len(df))],
columns = ["size","num"]
)
print(df['num'])
return df
fig = plt.figure(figsize=(9,5))
sns.set_style("darkgrid")
dfn = [extract(n) for n in range(1,10)]
df = pd.DataFrame({
f"S{i}" : df['num']
for i,df in enumerate(dfn)
})
df_melt = pd.melt(df)
print(df_melt.head())
#violinplot
sns.violinplot(x='variable', y='value', data=pd.melt(df), jitter=True)
plt.ylabel("Diameter(µm)")
fig.savefig("fig.png",dpi = 500)
>>>
Name: num, Length: 256, dtype: float64
variable value
0 S0 0.0
1 S0 0.0
2 S0 0.0
3 S0 0.0
4 S0 0.0
出力結果